PPIDM : mining protein-protein interactions to infer domain-domain interactions
What is PPIDM ?
PPIDM (Protein-Protein Interactions Domain Miner) is a computational method developed in 2019-2021 for inferring protein domain-domain interactions (DDIs) using multiple sources of protein-protein interactions (PPIs). Indeed, PPIs are physically mediated by DDIs. Structural databases (3DID, KBDOCK, …) contain validated DDIs extracted from 3D structures but these DDIs do not cover all PPIs of the interactome. Therefore, computational approaches are needed to infer DDIs from PPIs.
The PPIDM approach is an extension of our previously described « CODAC » (Computational Discovery of Direct Associations using Common neighbors) method for inferring new edges in a tripartite graph.
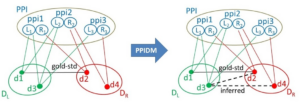
Schematic illustration of edge inference by PPIDM in a tripartite graph setting G (X, Y, Z, E). Z is here PPI, a set of ppis, X and Y are DL and DR, two sets of Pfam domains. Each item in PPI is an ordered pair of proteins ppi_i =(L_i, R_i) with Id(L_i) ≤ Id(R_i). Domains in DL and DR are connected to their common neighbor item ppi_i in PPI through L_i and R_i, the proteins they belong to, respectively. The (d1, d2) edge comes from the Gold-Standard dataset of DDIs. With PPIDM, new edges are inferred between domains of DL and domains of DR if their adjacency vectors in PPI are similar. Here, the (d3, d2)edge is inferred because d3 and d2 are found in ppi_1 and ppi_2, and (d3, d4) is inferred because d3 and d4 are found in ppi_2 and ppi_3. However, the score of (d3, d2) will be lower than the score of (d1, d2) because d3 has one neighbor that does not contain d2 (namely ppi3).
The PPIDM method has been applied to seven widely used PPI resources, using as « Gold-Standard » a set of DDIs extracted from 3D structural databases. Overall, PPIDM has produced a dataset of 84,552 non-redundant DDIs. Statistical significance (p-value) is calculated for each source of PPI and used to classify the PPIDM DDIs in Gold (9,175 DDIs), Silver (24,934 DDIs) and Bronze (50,443 DDIs) categories. Dataset comparison reveals that PPIDM has inferred from the 2017 releases of PPI sources about 46% of the DDIs present in the 2020 release of the 3did database, not counting the DDIs present in the Gold-Standard. The PPIDM dataset contains 10,229 DDIs that are consistent with more than 13,300 PPIs extracted from the IMEx database, and nearly 23,300 DDIs (27.5%) that are consistent with more than 214,000 human PPIs extracted from the STRING database. Examples of newly inferred DDIs covering more than 10 PPIs in the IMEx database are provided.
Further exploitation of the PPIDM DDI reservoir includes the inventory of possible partners of a protein of interest and characterization of protein interactions at the domain level in combination with other methods.
Availability
All code written in support of this publication is publicly available at
https://gitlab.inria.fr/capsid.public_codes/ppidmpublic.
Input files and generated data are available from Zenodo deposit at https://doi.org/10.
5281/zenodo.4880347.
Download a PPIDM dataset
The « gold » (highest confidence) DDIs can be uploaded here. The dataset (9175 DDIs) includes both gold-standard (2852) and inferred (6323) DDIs
Citing PPIDM
Alborzi SZ, Ahmed Nacer A, Najjar H, Ritchie DW, Devignes MD. (2021) PPIDomainMiner: Inferring domain-domain interactions from multiple sources of protein-protein interactions. PLoS Comput Biol. 17:e1008844. doi:10.1371/journal.pcbi.1008844
The approach is an extension of our previously described “CODAC” (Computational Discovery of Direct Associations using Common neighbors) method for inferring new edges in a tripartite graph, described in our GODM paper.
Alborzi SZ, Ritchie DW, Devignes MD (2018). Computational discovery of direct associations between GO terms and protein domains. BMC Bioinformatics.19:413. doi:10.1186/s12859-018-2380-2.
PPIdomainMiner was first described in Zia Alborzi’s European PhD Thesis (2018).
Acknowledgements
This work was carried out at the LORIA–Inria Nancy Grand-Est in the CAPSID team created by Dave Ritchie.
 Dave Ritchie , Inria Research Director, CAPSID team leader (2015-2019) passed away prematurely in September 2019. This work is dedicated to him, in memory of his interest in this work and in tribute to his many scientific contributions in structural bioinformatics.
Dave Ritchie , Inria Research Director, CAPSID team leader (2015-2019) passed away prematurely in September 2019. This work is dedicated to him, in memory of his interest in this work and in tribute to his many scientific contributions in structural bioinformatics.
 Seyed Ziaeddin Alborzi. Main designer and developer during his PhD Thesis in the CAPSID Team (2014-2018).
Seyed Ziaeddin Alborzi. Main designer and developer during his PhD Thesis in the CAPSID Team (2014-2018).
 Amina AHMED NACER, post-doc fellow in the CAPSID team, funded by CHRU Nancy and Région Lorraine in conjunction with MD DEVIGNES’s « contrat d’interface » (2019-2020).
Amina AHMED NACER, post-doc fellow in the CAPSID team, funded by CHRU Nancy and Région Lorraine in conjunction with MD DEVIGNES’s « contrat d’interface » (2019-2020).
 Hiba NAJJAR, student at the Ecole des Mines Nancy, intern in the CAPSID team in 2020.
Hiba NAJJAR, student at the Ecole des Mines Nancy, intern in the CAPSID team in 2020.
 Marie-Dominique DEVIGNES, CNRS Research associate, CAPSID team leader (2019-2024)
Marie-Dominique DEVIGNES, CNRS Research associate, CAPSID team leader (2019-2024)
Funding
- CPER IT2MP Innovation Technologiques et Modélisation pour la Médecine Personnalisée, Région Lorraine, Inria, Inserm, Université de Lorraine
- ANR PEPSI (ANR-11-MONU-006-02)
- ANR RHU FIGHT-HF (ANR-15-RHUS-0004)
- Cordi-S Inria PhD Fellowship
- Region Lorraine and Nancy Faculty Hospital post-doctoral fellowship
Contact
Marie-Dominique Devignes
Capsid Team – LORIA
Campus Scientifique
BP239
54500 Vandoeuvre-les-nancy cedex, France
marie-dominique.devignes AT loria.fr




